Prof Elizabeth (Lisa) Matisoo-Smith,
Professor of Biological Anthropology
Dept of Anatomy, School of Biomedical Sciences, University of Otago

What can DNA tell us about Pacific Origins and the Settlement of Aotearoa?
Elizabeth (Lisa) Matisoo-Smith, Professor of Biological Anthropology Dept of Anatomy, School of Biomedical Sciences, University of Otago
Elizabeth (Lisa) Matisoo-Smith is Professor of Biological Anthropology, and head of the Department of Anatomy of the University of Otago. She has published research on everything from rats in Russell to chickens in pre-Spanish South America, the Kiwi historical diet, the paper mulberry, the DNA of dogs, ancient Caribbean guinea pigs, African cattle, the genetics of Maronite Christians of Lebanon, the extermination of the Chatham Islands sea lion, and on following the mitochondria of rats and women throughout the peopling of the Pacific. In this paper she describes how genetics explains the movement of people and their plants and animals across the Pacific, and down to our island.
Where do we come from and how did we get here? This is a fundamental question that all peoples ask. Oral traditions and mythology often provide explanations, and more recently, academic scientific research has contributed evidence. From a western science perspective, we can try to identify population origins and track pathways of migration in a number of ways – traditionally, historians and others identified the similarities in languages and recognised that the languages of Polynesia were all very similar to each other, and were also similar to languages spoken in Island Southeast Asia. Archaeologists investigate the material culture of past societies and recognise similarities in artefact types, settlement patterns and other remains, which help them to reconstruct origins, trade patterns and other aspects related to human history. More recently, DNA evidence has been added to the package of scientific tools to reconstruct population origins and histories.
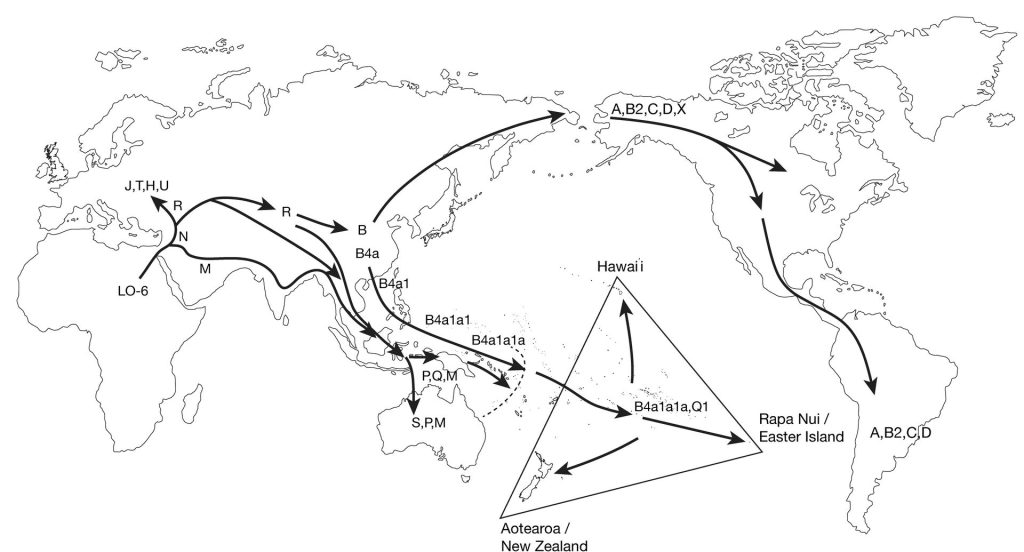
It was the ground-breaking work of New Zealand-born scientist, Allan Wilson, that really demonstrated to both scientists and the general public that we could use DNA to reconstruct human migration in the past. In particular, Wilson and his research group at the University of California, Berkeley, highlighted the value of a particular type of DNA, known as mitochondrial DNA, or mtDNA for short, for reconstructing population origins and migration pathways. MtDNA is inherited exclusively through the maternal line, so by identifying the maternal lineages of people from across the globe, we can reconstruct genetic relationships through time. In 1987, Wilson and his team showed, through analyses of mtDNA, that all modern human populations could be traced back to a common maternal ancestor, dubbed by the press, the Mitochondrial Eve, who lived in Africa sometime between 150,000 and 200,000 years ago. They also showed that the origins of all non-African populations could be traced back to a migration of a small group of people who left Africa about 60,000 to 65,000 years ago (Cann et al. 1987). As people moved, their mtDNA slowly changed and the maternal lineages diverged, or split to represent the large, diverse family tree we see today. Each branch of the mtDNA phylogeny, or family tree, is designated by a letter, A to Z. As each of those main branches split, we assign them a number, such as A1, A2 etc. When those branches split, we give each sub-branch another letter, so the A2 branch might split into A2a and A2b – and this process continues alternating letters and numbers to designate the new branches that grow from the original trunk of the tree – our last common shared ancestor.
The settlement of the Pacific, and in particular the settlement of Polynesia, is the result of one of the last major human migration events in history. Archaeologically, the settlement of Remote Oceania, or the region to the east of the Solomon Islands – including the earliest settlers in the Polynesian Triangle, is associated with what archaeologists have dubbed the Lapita culture. Lapita sites are most obviously identified by the remains of a distinct type of pottery with a set of design motifs that have been pressed into the pot with a comb-like tool. The process has been described as being similar and perhaps even representative of the tattooing of the pots with those specific design motifs – including many of the patterns that we see in traditional Polynesian tattoos today. Lapita archaeological sites first appear in the Pacific in the islands of the Bismarck Archipelago around 3,300 years ago. They very rapidly spread across the Pacific, reaching Vanuatu, New Caledonia, Fiji, Samoa and Tonga between 3000 and 2800 years ago (Kirch 2017). The settlement of the rest of the Polynesian Triangle did not happen for another 2000 years, with the earliest archaeological sites in central East Polynesia (the Cook Islands and the Society Islands – Tahiti, Raiatea etc) dating to about 1200 years ago. Finally, Hawaii, Rapa Nui Easter Island and Aotearoa New Zealand were settled between 1000 and 750 years ago (Wilmshurst et al. 2011). All Polynesian languages and most of the others of Remote Oceania, which also includes most of the languages of Micronesia, belong to a language family known as the Austronesian languages. The homeland or origin of the Austronesian languages has been identified as Taiwan. These were the languages that were spoken by the indigenous Taiwanese peoples before the island was colonised by the Han Chinese.
Many studies have also investigated the origins of Pacific peoples through analyses of the mtDNA. Polynesian peoples have maternal lineages that almost exclusively belong to the B branch of the mtDNA family tree (unless someone has a non-Polynesian maternal ancestor). In fact, the majority of Polynesians, including almost all NZ Māori with strict Māori maternal ancestors, have mtDNA lineages that belong on a distant sub-branch of B, known as B4a1a1 or perhaps another branch that is descended from that lineage (e.g. B4a1a1c or B4a1a1m). When we look at the distribution of the B4a1a1 lineage, we see that it is found almost exclusively in Pacific populations. Its immediate ancestral lineage B4a1a is found in populations within Island Southeast Asia. The B lineage originated in East Asia about 50,000 years ago, and from there we can trace it back to the common maternal ancestor that left Africa and ultimately to the “Mitochondrial Eve”.
Because there is not a huge amount of mtDNA variation within Polynesian populations, it is difficult to track Māori origins beyond a broad general region in Central East Polynesia. All of the mtDNA lineages we see in Māori are also found in the Cook Islands, the Society Islands and elsewhere in East Polynesia. We can also see most of these in West Polynesian populations, in Samoa and Tonga for example. The mtDNA linking of Māori origins to the Cook Islands and Society Islands also fits with the linguistic and archaeological data as well as the oral traditions. This provides some scientific evidence for the location of the East Polynesian homeland region, often referred to as “Hawaiiki” in mythology (Walter et al. 2017). Linguistically, we also see the links of these East Polynesian languages to those of Samoa and Tonga, and from there through the western Pacific. These “Oceanic” languages, which are a subgroup of Austronesian languages, have a distribution in the western Pacific that correlates closely, though not exclusively, with the location of Lapita archaeological sites, and so Austronesian origins, and therefore Māori and Polynesian origins, have often been linked genetically, archaeologically and linguistically to the Austronesian migration through Island Southeast Asia and ultimately to East Asia and then Africa.
In an attempt to try to tease apart the history and relationships between the closely related populations of East Polynesia and Pacific populations more broadly, it was recognised that there might be an alternative approach, again, using mtDNA. It was recognised that Pacific peoples since the Lapita period carried a number of plant and animal species with them in their canoes as they settled the islands across the Pacific. This “transported landscape” was one of the ways that Pacific peoples adapted to the range of new environments they encountered and is also perhaps why they were so successful in establishing those new communities on the islands across the Pacific. We realised that if we could study the mtDNA of the animals that people carried in their colonising canoes, and reconstruct the dispersal histories of those animals, then we would be able to tell where those canoes came from and thus could identify the origins and interactions of the people who sailed those canoes. This approach was called the “commensal model” for tracing human origins and mobility. The commensal model was first applied to tracking the origins and dispersal of the Pacific rat, or kiore (Rattus exulans) (Matisoo-Smith 1994).
The kiore was chosen as the first animal to use for this new approach for several reasons, but primarily because it is the only animal carried by the Polynesians and their ancestors that is a distinct species from the animals transported by later European explorers and colonists. The domesticated pigs and chickens that Lapita peoples introduced in their colonising canoes and the dogs introduced later and distributed throughout Polynesia belong the same species of pigs, dogs and chickens as those introduced later by Europeans, so they have interbred and the DNA lineages we see in the Pacific today are European lineages. The kiore, on the other hand, are a distinct species from the rats introduced by Europeans (Rattus rattus and Rattus norvegicus, the ship rat and Norway rat). Because they are different species, they cannot interbreed – therefore the kiore we see on islands today are the direct descendants of those originally introduced by Pacific people hundreds if not thousands of years ago. The skeletal remains of kiore are found in the earliest archaeological sites across the Lapita distribution and throughout Polynesia. The distribution of the pigs, chickens and dogs (kuri) in the Pacific are much patchier, for example, only the kiore and kuri were successfully introduced to Aotearoa by Māori. Oral traditions tell us that kiore were an important food source for many Pacific peoples, including in Aotearoa (see https://teara.govt.nz/en/video/16131/eating-kiore ). It was through talking with kaumātua like Hori Parata (Ngāti Wai) and colleagues like Dr Mere Roberts, who shared their knowledge of the importance of kiore in Māori and Polynesian society and thus their value for reconstructing the origins of the waka that brought them, that allowed me to recognise the opportunity and develop the commensal model.
As part of my PhD research at the University of Auckland, I undertook a study of mtDNA diversity in Polynesian kiore populations. We found that the kiore from Aotearoa were most closely related to kiore from the Cook and the Society Islands – with evidence of a southern network incorporating these islands with the Kermadec Islands and Aotearoa. In addition, we found a northern Polynesian network incorporating the Society Islands, the Marquesas Islands and Hawai’i (Matisoo-Smith et al. 1998). Later, as the technology developed that allowed us to obtain mtDNA from archaeological remains, we were able to not only confirm that the modern kiore populations were indeed directly descended from the ancient kiore populations. We were also able to track the origins of kiore through the rest of the Pacific and into Island Southeast Asia. We found that there were actually several interaction spheres throughout the Pacific, and the Polynesian kiore could be traced back to the island of Halmahera in the Maluku Islands (Matisoo-Smith and Robins 2004). We also realised that we could use these ancient DNA (aDNA) methods to study the archaeological remains from other introduced animals, such as kuri, pigs and chickens (Matisoo-Smith 2007).
Over the last 20 years these aDNA studies have shown that the origins and dispersal history of each of these species are unique, and so together this provides even more evidence of the complexity of human history in the Pacific. The ancient Pacific pig DNA indicates an origin on mainland SE Asia, near Vietnam. Only a single lineage of pigs was introduced to the Pacific and these were transported through the more western islands of Indonesia to Papua New Guinea and into the Pacific with Lapita peoples (Larson et al. 2007). Multiple mtDNA lineages of dogs were introduced into the Pacific, at a number of different times, but dogs were not part of the Lapita introductions, arriving in Polynesia at about the time that expansion into Central East Polynesia begins (Greig et al. 2018; Greig et al. 2016). The ancient chicken DNA studies show that chickens were also introduced by Lapita people, but sometime after 1500 years ago, a second mtDNA lineage of chickens was introduced. We also found evidence of Polynesian contact with South America by 1350 AD (Storey et al. 2013). This evidence of South American contact by Polynesians is also supported by linguistic, archaeological and genetic data which all indicate that Polynesian voyagers picked up the kumara, probably in Ecuador, and brought it back to Central East Polynesia in time for it to be dispersed to Hawai’i, Rapa Nui and Aotearoa (Green 2005; Hather and Kirch 1991; Roullier et al. 2013). Ancient DNA techniques have also been applied to study the origins and dispersal pathways of the paper mulberry plant (Broussonetia papyrifera), which is used to make bark cloth, or tapa, throughout Island Southeast Asia and the Pacific (Olivares et al. 2019). This work indicates that the Pacific plants originate in Taiwan and that there were numerous dispersal networks and likely interaction spheres within the Pacific – with later, historic introductions of the plants to Hawai’i, most likely with migrant workers from China and Japan.
Ancient DNA methods and technology are now also being applied to ancient human remains, or kōiwi tangata. The archaeological site of Wairau Bar, located at the top of the South Island at the mouth of the Wairau River, and dated to the early 1300s, is one of Aotearoa’s earliest sites and one which has provided a large amount of data regarding the initial settlement of Aotearoa. A large amount of cultural material, moa bones and eggs as well as other large fauna, and the remains of over 42 burials were recovered from the site, initially by fossickers and then, later, as a result of excavations by the Canterbury Museum from the 1940s through the 1960s. This material was all stored and much of it displayed at the Canterbury Museum. In 2008 discussions began between Rangitāne, the iwi with kaitiakitanga status at Wairau Bar, and the museum regarding repatriation of the tūpuna and associated artifacts. Researchers from the University of Otago were brought into these discussions to advise on the types of scientific analyses that might be possible prior to reburial of the skeletal remains of the tūpuna. It was determined that a small archaeological survey and excavation could be undertaken to locate an appropriate urupā and that a full biological study could be undertaken on the kōiwi – it was also agreed that some aDNA and other destructive analyses could be undertaken on a small number of samples to investigate origins, diet and, particularly, if those studies might help to address and better understand some of the health issues facing Māori and Pacific peoples today (Brooks et al. 2009).
The DNA and other studies of the Wairau Bar tūpuna began soon after the memorandum of understanding was signed between the Canterbury Museum, Rangitāne and the University of Otago. Ancient mtDNA was obtained from several of the burials and we found the same mtDNA lineages seen in Māori and other East Polynesian populations today (Knapp et al. 2012). The numbers of different B4a1a lineages identified indicates a large founding population that was not closely maternally related. Estimates are likely to be in the range of several hundreds of people if not more, again, indicated a mass migration from the Central Polynesian Hawaiiki. Isotope studies indicated that some of the burials, likely spent their childhood in a foreign location – very possibly in Central East Polynesia (Kinaston et al. 2013). A full assessment of the bones indicated that the populations was generally healthy, but there were some evidence of childhood stress and possible infectious and non-infectious disease (Buckley et al. 2010). Specifically, there were lesions on the bones that indicate several individuals suffered from gout and possibly diabetes or other metabolic diseases. Similar lesions have been identified in other ancient Pacific remains in both Lapita associated burials in Vanuatu and in CHamoru burials from Guam, and peoples across the Pacific today all suffer very high rates of metabolic disease like gout and diabetes (Buckley 2011).
Based on this skeletal evidence suggesting that metabolic disease in the Pacific has a very deep history and is therefore not solely the result of an adaptation to western lifestyles and lack of exercise, which has often been suggested, we are now investigating the genetic history of gout and metabolic disease in modern Pacific (Gosling et al. 2015; Gosling et al. 2014a; Gosling et al. 2014b). Studies of modern populations in the Pacific indicate that while rates of gout and diabetes are high throughout the Pacific, East Polynesians and West Polynesians have different genetic markers that are associated with the high rates of metabolic disease (Phipps-Green et al. 2010). When we combine this data with the archaeological evidence and aDNA evidence of the faunal remains from across the Pacific, we think that perhaps the immediate origins of East Polynesian peoples are not exclusively within West Polynesia. There are several indicators of possible influences from or connections to Micronesia, and we have suggested that this may be evidence for the arrival of a new group of people in the Pacific who passed from Island Southeast Asia through the atolls of Micronesia. This may have been a factor in the timing of the final pulse of peoples that resulted in the settlement of Central East Polynesia around 1200 years ago (Addison and Matisoo-Smith 2010). We also think that the genetic markers that have been found in Pacific peoples that appear to be associated with gout may have actually provided the ancestors with an advantage for adapting to new environments, with gout possibly being associated with better chances of survival from malarial infection (Matisoo-Smith and Gosling 2018). Malaria is present both in Island Southeast Asia and in the Western Pacific region that the Lapita ancestors presumably passed through, so it could have impacted the ancestors of all Pacific peoples, even though malaria has never been present in Polynesia or Micronesia.
In conclusion, DNA data is providing us with a much better understanding of the population origins and settlement histories in the Pacific, including Aotearoa. It is becoming increasingly clear that it was a very complex process or processes, with evidence of strategic planning, significant levels of mobility and interaction, by people who were highly successful adapting behaviourally and technologically to a range of environments. We also see that multidisciplinary research is necessary to unravel this complex history. Researchers from a range of fields need to work together and collaborate, but perhaps more importantly, we need the engagement of indigenous communities to both highlight their own concerns and questions for and about research and to contribute their own knowledge to the design and interpretation of data generated by academic research. By walking together, and learning from the past, we can move more confidently and positively into the future.
References
Addison DJ, and Matisoo-Smith E. 2010. Rethinking Polynesian Origins: A West-Polynesian Triple-I Model. Archaeology in Oceania 45:1-12.
Brooks E, Jacomb C, and Walter R. 2009. Archaeological investigations at Wairau Bar. Archaeology in New Zealand 52(4):259-268.
Buckley H, Tayles N, Halcrow SE, Robb K, and Fyfe R. 2010. The people of Wairau Bar: A re-examination. Journal of Pacific Archaeology 1(1):1-20.
Buckley HR. 2011. Epidemiology of Gout: Perspectives from the Past. Current Rheumatology Reviews 7:106-113.
Cann RL, Stoneking M, and Wilson AC. 1987. Mitochondrial DNA and human evolution. Nature 325:31-36.
Gosling AL, Buckley HR, Matisoo-Smith E, and Merriman TR. 2015. Pacific Populations, Metabolic Disease and ‘Just-So Stories’: A Critique of the ‘Thrifty Genotype’ Hypothesis in Oceania. Annals of Human Genetics 79(6):470-480.
Gosling AL, Matisoo-Smith E, and Merriman TR. 2014a. Gout in Māori. Rheumatology 53(5):773-774.
Gosling AL, Matisoo-Smith E, and Merriman TR. 2014b. Hyperuricaemia in the Pacific: why the elevated serum urate levels? Rheumatology International 34(6):743-757.
Green RC. 2005. Sweet potato transfers in Polynesian prehistory. In: Ballard C, Brown P, Bourke RM, and Harwood T, editors. The Sweet Potato in Oceania: A Reappraisal. Sydney: Oceania Publications. p 43-62.
Greig K, Gosling AL, Collins C, Boocock J, McDonald K, Addison DJ, Allen MS, David B, Gibbs M, Higham CFW et al. . 2018. Tracking dogs across the Pacific: ancient mitogenomes reveal a complex history of origins and translocations. Scientific Reports 8:9130.
Greig K, Walter R, and Matisoo-Smith EA. 2016. Dogs and people in Southeast Asia and the Pacific. In: Routledge, editor. The Routledge Handbook of Bioarchaeology in Southeast Asia and the Pacific Islands. p 462-482.
Hather J, and Kirch PV. 1991. Prehistoric sweet potato (Ipomoea batatas) from Mangaia Island, Central Polynesia. Antiquity 65:887-893.
Kinaston RL, Walter RK, Jacomb C, Brooks E, Tayles N, Halcrow SE, Stirling C, Reid M, Gray AR, Spinks J et al. . 2013. The First New Zealanders: Patterns of Diet and Mobility Revealed through Isotope Analysis. PLoS One 8(5):e64580.
Kirch PV. 2017. On the Road of the Winds: An Archaeological History of the Pacific Islands before European Contact. Berkeley, CA: University of California Press.
Knapp M, Horsburgh KA, Prost S, Stanton J-A, Buckley HR, Walter RK, and Matisoo-Smith EA. 2012. Complete mitochondrial DNA genome sequences from the first New Zealanders. Proceedings of the National Academy of Sciences 109(45):18350-18354.
Larson G, Cucchi T, Fujita M, Matisoo-Smith E, Robins J, Anderson A, Rolett B, Spriggs M, Dolman G, Kim T-H et al. . 2007. Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proceedings of the National Academy of Sciences of the United States of America 104(12):4834-4839.
Matisoo-Smith E. 1994. The Human Colonisation of Polynesia. A Novel Approach: Genetic Analyses of the Polynesian Rat (Rattus exulans). J Polynesian Soc 103:75-87.
Matisoo-Smith E. 2007. Animal translocations, genetic variation and the human settlement of the Pacific. In: Friedlaender JS, editor. Genes, Language and Culture History in the Southwest Pacific. Oxford: Oxford University Press. p 157–170.
Matisoo-Smith E, and Gosling AL. 2018. Walking backwards into the future: the need for a holistic evolutionary approach in Pacific health research. Annals of Human Biology 45(3):175-187.
Matisoo-Smith E, Roberts RM, Irwin GJ, Allen JS, Penny D, and Lambert DM. 1998. Patterns of prehistoric human mobility in Polynesia indicated by mtDNA from the Pacific rat. Proceedings of the National Academy of Sciences of the United States of America 95(25):15145-15150.
Matisoo-Smith E, and Robins JH. 2004. Origins and dispersals of Pacific peoples: Evidence from mtDNA phylogenies of the Pacific rat. Proceedings of the National Academy of Sciences of the United States of America 101(24):9167-9172.
Olivares G, Peña-Ahumada B, Peñailillo J, Payacán C, Moncada X, Saldarriaga-Córdoba M, Matisoo-Smith E, Chung K-F, Seelenfreund D, and Seelenfreund A. 2019. Human mediated translocation of Pacific paper mulberry [Broussonetia papyrifera (L.) L’Hér. ex Vent. (Moraceae)]: Genetic evidence of dispersal routes in Remote Oceania. PLOS ONE 14(6):e0217107.
Phipps-Green AJ, Hollis-Moffatt JE, Dalbeth N, Merriman ME, Topless R, Gow PJ, Harrison AA, Highton J, Jones PBB, Stamp LK et al. . 2010. A strong role for the ABCG2 gene in susceptibility to gout in New Zealand Pacific Island and Caucasian, but not Māori, case and control sample sets. Human Molecular Genetics 19(24):4813-4819.
Roullier C, Benoit L, McKey DB, and Lebot V. 2013. Historical collections reveal patterns of diffusion of sweet potato in Oceania obscured by modern plant movements and recombination. Proceedings of the National Academy of Sciences 110(6):2205-2210.
Storey AA, Quiroz D, Beavan N, and Matisoo-Smith E. 2013. Polynesian chickens in the New World: a detailed application of a commensal approach. Archaeology in Oceania 48:101-119.
Walter R, Buckley H, Jacomb C, and Matisoo-Smith E. 2017. Mass Migration and the Polynesian Settlement of New Zealand. Journal of World Prehistory 30(4):351-376.
Wilmshurst JM, Hunt TL, Lipo CP, and Anderson AJ. 2011. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proceedings of the National Academy of Sciences 108(5):1815-1820.
Recommended Reading – the following papers and books provide an overview or extended information and are written for a general audience:
Jones TL, Storey AA, Matisoo-Smith E, and Ramirez-Aliaga JM, editors. 2011. Polynesians in America: Pre-Columbian contacts with the New World. Plymouth: AltaMira Press.
Kirch PV. 2017. On the Road of the Winds: An Archaeological History of the Pacific Islands before European Contact. Berkeley, CA: University of California Press.
Matisoo-Smith E. 1994. The Human Colonisation of Polynesia. A Novel Approach: Genetic Analyses of the Polynesian Rat (Rattus exulans). J Polynesian Soc 103:75-87.
Matisoo-Smith E, and Daugherty C. 2012. Africa to Aotearoa: the longest migration. Journal of the Royal Society of New Zealand 42(2):87-92.
Matisoo-Smith E, and Horsburgh KA. 2012. DNA for archaeologists. Walnut Creek, Calif.:
Mitochondrial DNA Tree
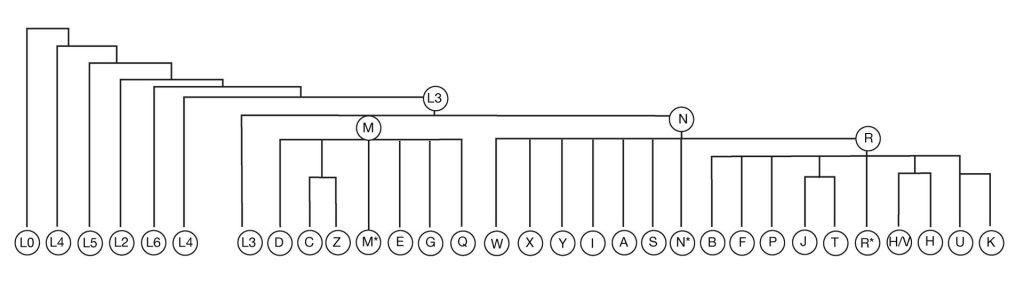
Figure 2 - Map